 ¶
¶
These docs are for iminuit version: 2.25.2 compiled with ROOT-v6-25-02-7198-g503faf859b





iminuit is a Jupyter-friendly Python interface for the Minuit2 C++ library maintained by CERN’s ROOT team.
Minuit was designed to optimize statistical cost functions, for maximum-likelihood and least-squares fits. It provides the best-fit parameters and error estimates from likelihood profile analysis.
The iminuit package brings additional features:
Builtin cost functions for statistical fits to N-dimensional data
Unbinned and binned maximum-likelihood + extended versions
Least-squares (optionally robust to outliers)
Gaussian penalty terms for parameters
Cost functions can be combined by adding them:
total_cost = cost_1 + cost_2Visualization of the fit in Jupyter notebooks
Support for SciPy minimizers as alternatives to Minuit’s MIGRAD algorithm (optional)
Support for Numba accelerated functions (optional)
Minimal dependencies¶
iminuit is promised to remain a lean package which only depends on numpy, but additional features are enabled if the following optional packages are installed.
numba: Cost functions are partially JIT-compiled ifnumbais installed.matplotlib: Visualization of fitted model for builtin cost functionsipywidgets: Interactive fitting, see example below (also requiresmatplotlib)scipy: Compute Minos intervals for arbitrary confidence levelsunicodeitplus: Render names of model parameters in simple LaTeX as Unicode
Documentation¶
Checkout our large and comprehensive list of tutorials that take you all the way from beginner to power user. For help and how-to questions, please use the discussions on GitHub or gitter.
Lecture by Glen Cowan
In the exercises to his lecture for the KMISchool 2022, Glen Cowan shows how to solve statistical problems in Python with iminuit. You can find the lectures and exercises on the Github page, which covers both frequentist and Bayesian methods.
Glen Cowan is a known for his papers and international lectures on statistics in particle physics, as a member of the Particle Data Group, and as author of the popular book Statistical Data Analysis.
In a nutshell¶
iminuit can be used with a user-provided cost functions in form of a negative log-likelihood function or least-squares function. Standard functions are included in iminuit.cost, so you don’t have to write them yourself. The following example shows how to perform an unbinned maximum likelihood fit.
import numpy as np
from iminuit import Minuit
from iminuit.cost import UnbinnedNLL
from scipy.stats import norm
x = norm.rvs(size=1000, random_state=1)
def pdf(x, mu, sigma):
return norm.pdf(x, mu, sigma)
# Negative unbinned log-likelihood, you can write your own
cost = UnbinnedNLL(x, pdf)
m = Minuit(cost, mu=0, sigma=1)
m.limits["sigma"] = (0, np.inf)
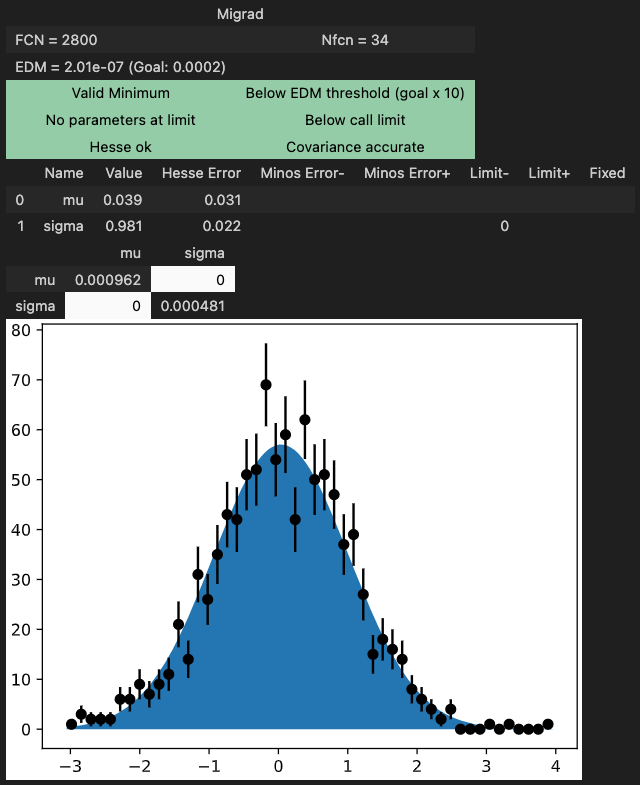
m.migrad() # find minimum
m.hesse() # compute uncertainties
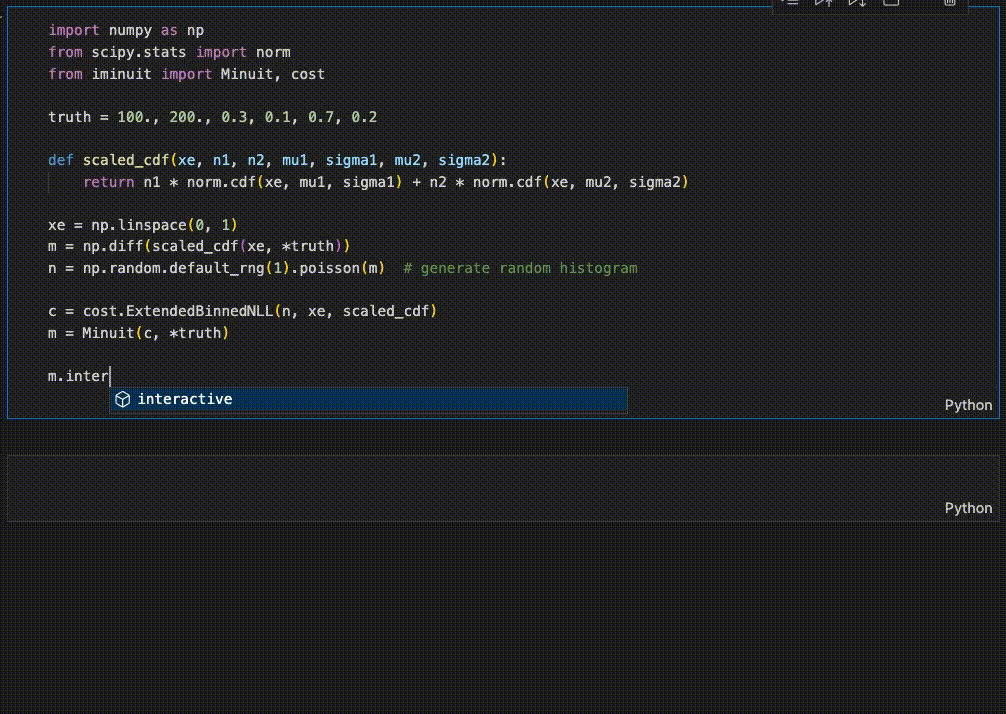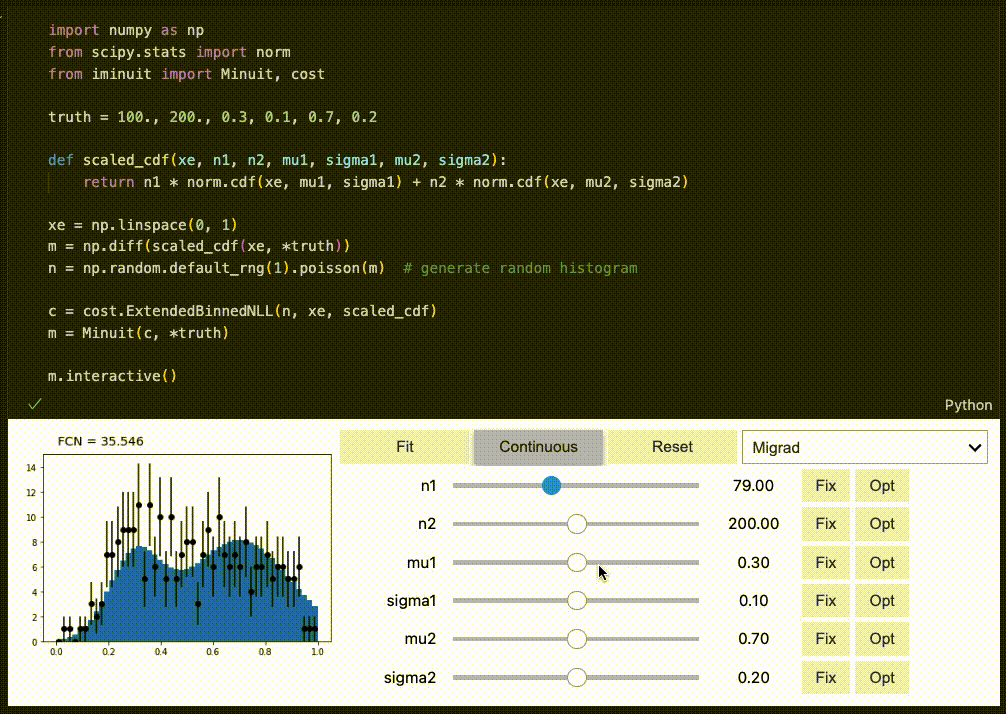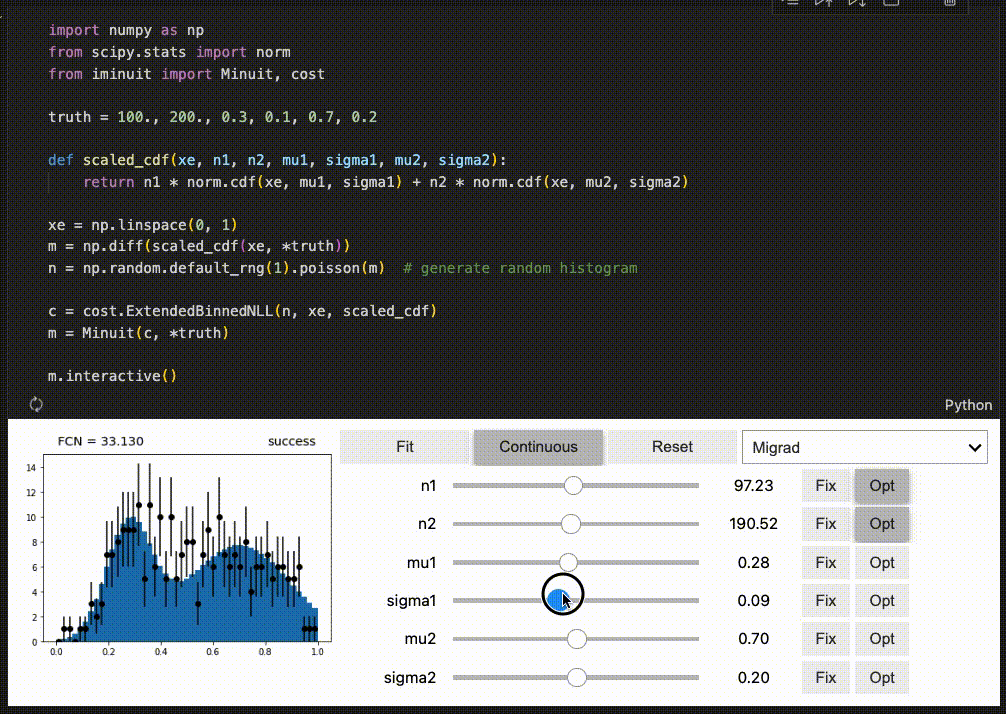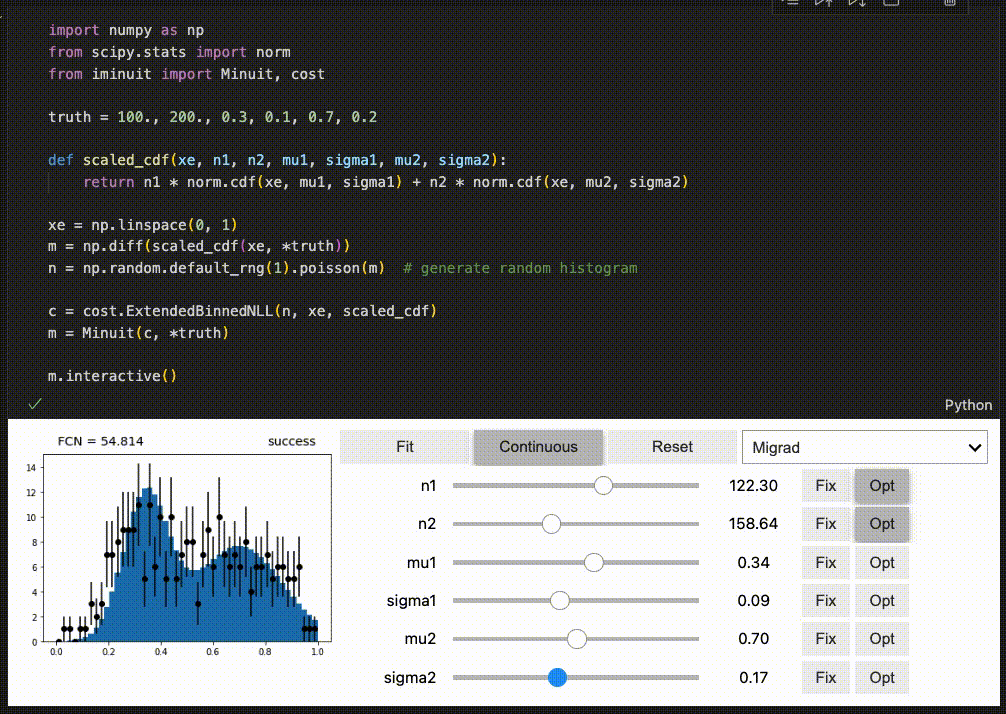
Interactive fitting¶
iminuit optionally supports an interactive fitting mode in Jupyter notebooks.
High performance when combined with numba¶
When iminuit is used with cost functions that are JIT-compiled with numba (JIT-compiled pdfs are provided by numba_stats ), the speed is comparable to RooFit with the fastest backend. numba with auto-parallelization is considerably faster than the parallel computation in RooFit.
More information about this benchmark is given in the Benchmark section of the documentation.
Partner projects¶
numba_stats provides faster implementations of probability density functions than scipy, and a few specific ones used in particle physics that are not in scipy.
boost-histogram from Scikit-HEP provides fast generalized histograms that you can use with the builtin cost functions.
jacobi provides a robust, fast, and accurate calculation of the Jacobi matrix of any transformation function and building a function for generic error propagation.
Versions¶
The current 2.x series has introduced breaking interfaces changes with respect to the 1.x series.
All interface changes are documented in the changelog with recommendations how to upgrade. To keep existing scripts running, pin your major iminuit version to <2, i.e. pip install 'iminuit<2' installs the 1.x series.